Home > Teams > Infectious Diseases with special emphasis on Co-Morbidities > HCEMM-BRC Pharmacodynamic Drug Interaction Research Group
GROUP LEADER: Viktória Lázár, Ph.D.
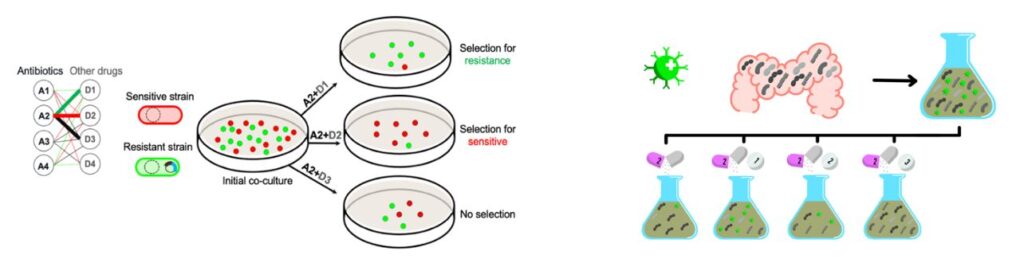
While antibiotics are the main weapons to fight and clear bacterial infection, they can also select for resistant pathogens and disrupt the gut microbiota. Although these ‘collateral damages’ are recognized, we currently lack treatment strategies to break the vicious cycle of antibiotic resistance while protecting the integrity of the host microbiome. By systematically screening drug interactions between traditional antibiotics and non-antibiotic pharmaceuticals (such as antivirals, vitamins, steroids, immunosuppressants, antidiabetic agents, anticoagulants etc.) the team is looking for novel drug combinations that can be used to clear resistant pathogens orders of magnitude more efficiently than its susceptible counterpart and maximize pathogen elimination while protecting the healthy gut-microbiota. Her HCEMM research integrates multidrug screens, genomic sequencing, laboratory evolution experiments, metagenomics, and cultivation-basedphenotyping of individual-specific microbiomes to discover optimal treatment strategies of high-risk infections for patients with chronic diseases.
The early diagnosis of the onset of chronic and infectious diseases is essential for their successful treatment. We are working on the development of non-invasive tests for the early detection of diseases such as cancers, sepsis or viral infections using molecular markers, which are present in the blood stream of patients. Circulating Nucleic Acids (CNA), which are packaged into exosomes can be used as a target for the development of qPCR assays, which can be performed routinely at the clinics or in blood laboratories. It is our goal to identify molecular markers, which are informative of the presence of early disease stages and work with industrial partners to create tests, which can either be used during an annual check up or when a patient is suspected of developing the condition.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 739593. HCEMM supported by EU Programme: H2020-EU.4.a. – Teaming of excellent research institutions and low performing RDI regions. Project starting date was 1 April 2017.






